Automancer
Automancer is a software application which makes it possible to automate lab protocols.
Automancer is written in Python 3.11 for the backend and TypeScript for the frontend.
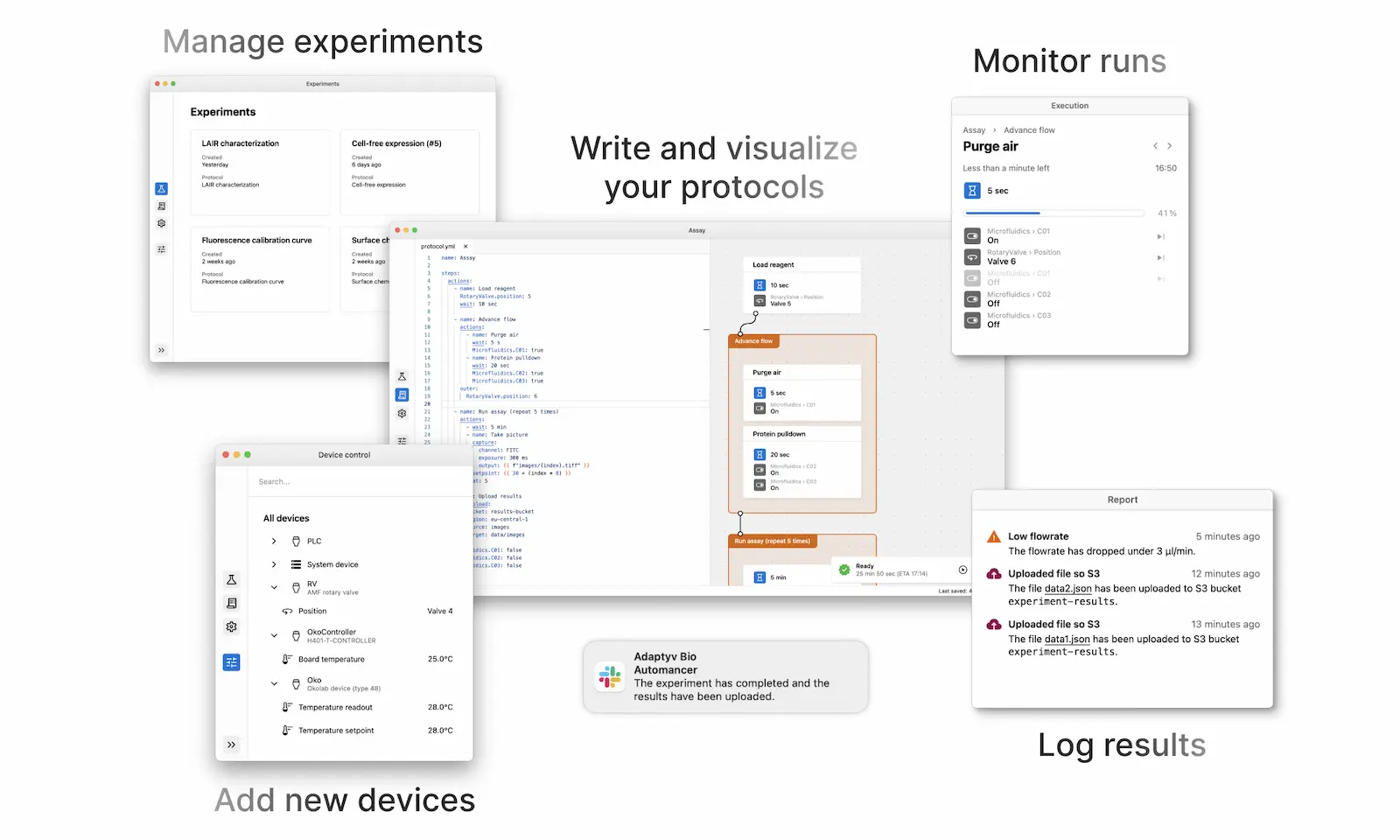
Features
- Modularity – Apart from the core functionality and UI, most of Automancer's capabilities are provided through plugins. Plugins are Python modules that can extend Automancer in one or more ways: they can add support for a new device, provide a custom protocol logic feature, report information to the UI… pretty much whatever you can dream up.
- User interface – Automancer's user interface provides easy control to users with no coding expertise. Furthermore, by supporting plugin development, we make extending the user interface simple, in particular when employing the built-in UI components. For instance, this can be used to report data generated through Python, allowing Python developers without a background in UI development to customize their view of the application.
- Protocol format – The declarative and human-readable (YAML-like) text format used by Automancer has numerous benefits. Unlike proprietary formats, text files are lightweight, easy to share, to reuse and to compare between iterations. The language can be learned quickly by beginners and does not require any coding expertise. Experienced users can extend the language using plugins. Automancer provides an optional built-in editor with advanced programmatic language features, such as live completion, errors and documentation on hover.
- Remote control – Experiments can be started and controlled from another computer on the same or another network connected to the Internet. This allows multiple people in different locations to monitor the same experiments, or when running experiments on multiple setups, to control them all from the same computer.
- Written in Python and open source – Automancer is written in Python for the backend and TypeScript/SCSS with React for the user interface, running on Electron. It is available under the MIT license and open to contributions.
Downloads
- macOS 11 and later – Intel, Apple Silicon using Rosetta 2
- Windows 10 and 11
Contents
Start by following the Getting started guide.